Users need to submit sequence (a cDNA or transcript) as target for siRNA design. A typical siRNA target sequence can be a cDNA, EST, Unigene, mRNA or transcript of gene from genomic sequencing project,
etc. The server will design potential siRNAs on the basis of user submission. Prior to analysis, back-end pipeline will check users submission by the following standards:
- A valid sequence can only be FASTA format or single sequence without FASTA header(Pure Sequence, see above figures);
- Server only allow users to submit one sequence for analysis once and maximal submission size is 15K;
- The submitted sequence should be between 30 and 15360 nt in length, and pipeline will report error for invalid submission;
- Only 'ATCGUN' are valid sequence letters, other characters will be deleted or changed to 'N'.
Choose the species name in which design siRNA will use to silence users’ submitted sequence. Selecting the species can automatic load its cDNA/transcript libraries for the assessment of genome wide off-target of siRNAs.
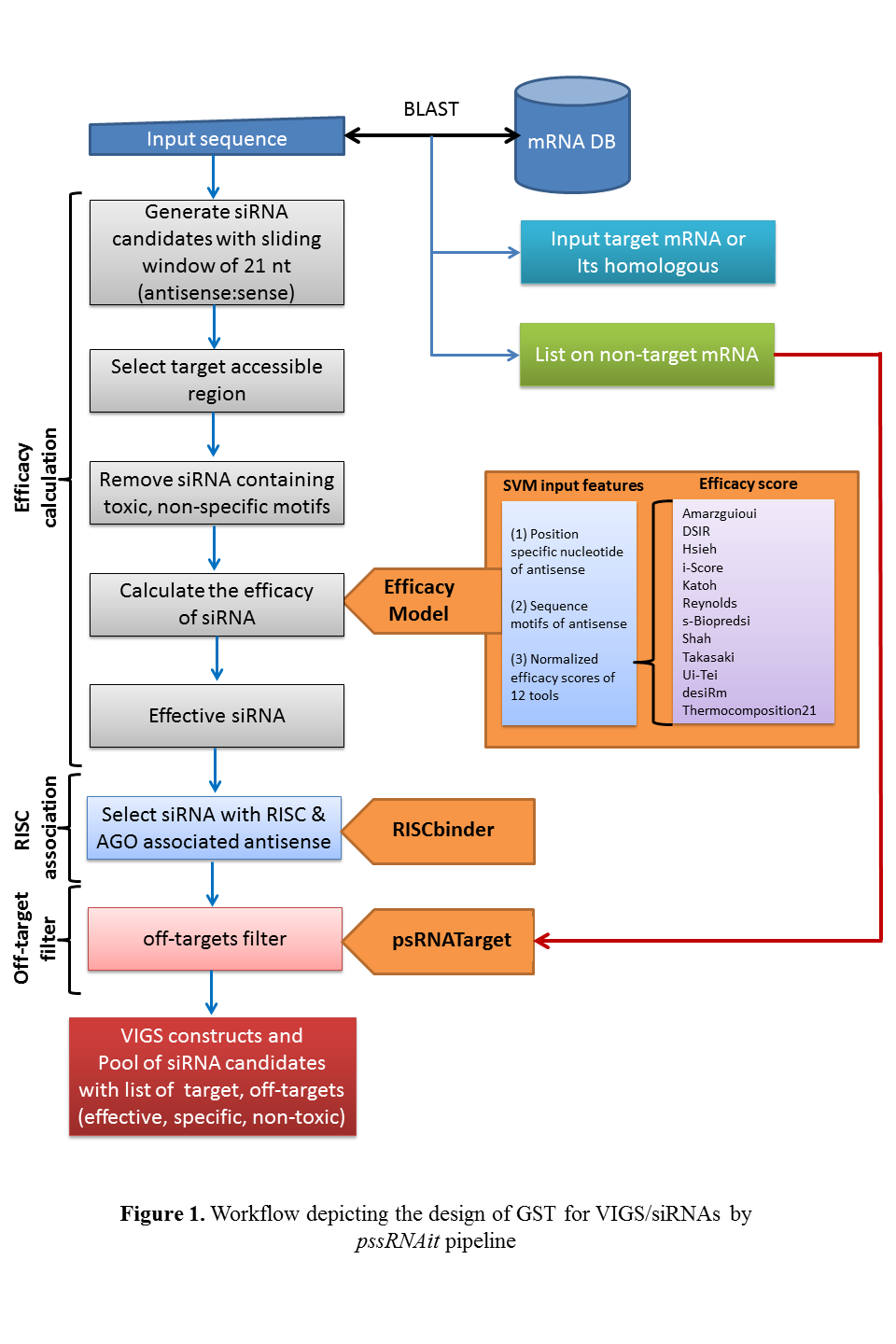
siRNA should be removed which contains toxic motifs. Studies showed that sequence motifs UGUGU, GUCCUUCAA (1,2), or UGGC (3) in siRNA show immunostimulatory activity and resulting reduce cell viability in the transfected animal cells (1,4-6). Moreover, some sequence motifs are commonly present among several genes. Therefore, it must avoid targeting those common motif otherwise antisense leads to silencing non-specific targets (7,8). Thus, we avoid uninterrupted >2x(CUG) (7,8), >2x(CCG), 2x(CGG) (9,10) repeat or WUAAAUW (11) motifs in the antisense strand of animal siRNA. We also avoid contiguous motif of >2x(CAN), or repeat of more than 4 same nucleotides like, AAAA, CCCC, GGGG, UUUU in siRNA of both animals and plants.
siRNA efficiency: Efficiency denotes the effectiveness of designed siRNA to silence the users’ submitted transcripts. The efficiency range can vary from 0-10, higher the value greater silencing of submitted transcript, eg: value 0 indicates no silencing while value 10 indicates 100% silencing. The default cut-off efficacy is 6 which display all siRNA whose efficacy is equal or more than 6. Users are advised to increase efficiency to get display of highly effective siRNA.
Maximum # of off-targets: In order to get specific siRNA, there should be no off-target. Specific siRNA can only silence users’ submitted transcripts without affecting to other genes. However, practically it is not possible to get every siRNA without off-target. Therefore, users can minimize the maximum number of off-target, default is 20, to find those siRNA which is specific and cause less off-target silencing.
Expect: Expect is the score to denotes complementarity between siRNA and off-targets transcripts based upon scoring schema of psRNATarget web server (http://plantgrn.noble.org/psRNATarget/) derived from small properties of no-coding RNA:target gene analyzed in the plant (12). Roughly, lower Expectation denotes more complementarity between siRNA:off-targets and chances are high for off-target silencing. The default cut-off threshold is 5 and will be include all siRNA:off-target whose score is in the range of 0-5. More stringent cut-off threshold [0-2.0] can’t find all possible off-target however more relaxed cut-off threshold [4.0-5.0] result higher prediction coverage of off-targets.
Off-target accessibility (UPE): In past, several investigations reported that target site should be accessibility in mRNA to bind with siRNA (13,14). Due to secondary structure in mRNA, some target site is inaccessible to interact with siRNA resulting no silencing. The RNAup was employed to calculate target accessibility, which is represented by the energy required to open secondary structure around target site (15). The less energy means the more possibility that siRNA is able to contact (and cleave) with mRNA and is represented by UPE. UPE can vary from 0-100 representing accessible-inaccessible, respectively. The default value is 25 which display all siRNA which have UPE in the range of 0-25.
It display the name of cDNA/transcripts which show nearly complete homology with users’ submitted sequence with users’ selected species specific cDNA/transcript libraries. These homologous sequences are excluded from off-target genes analysis because: either homolog sequence is same with users’ submitted sequence, or homolog sequence is the isoforms of users’ submitted sequence. However, users can deselect the homologous transcripts if want to consider them as off-targets and resubmit by click the query button to get potent siRNA and off-targets.
RISCbinding antisense and sense score can vary from negative to positive,
where more negative value indicates higher probability of siRNA strand not to load into RISC, while more positive value
indicates higher probability of siRNA strand to load into RISC complex. Users need to select higher RISCbinding score for antisense than that of sense
stand so that antisense can sort into RISC complex for gene silencing.
VIGS candidates could be customized using following option.
Range of VIGS length: At default, length of VIGS is 100-300 nt, however, user can modify it.
Minimal # of siRNAs in VIGS candidates: At default, minimum number of siRNA in a VIGS must have 4 siRNA, however, user can modify it.
Minimal distance of two effective siRNAs: At default, minimum distance between two siRNA is 10 nt., however, user can increase the distance between two siRNA.
Argonaute (AGO) is the catalytic engine of RISC complex. In plants, the sorting of siRNA into different Argonaute (AGO) protein is mainly determined by 5’-terminaus of siRNA (Mi, Cai et al. 2008).).
Features of AGO proteins in Arabidopsis thaliana (Fang and Qi 2016).
AGO1: bind with miRNAs or ta-siRNAs[1] and cleave the target mRNA.
AGO2: binds with siRNA
AGO4: binds with hc-siRNAs[2] and ta-siRNAs and involve in DNA methylation and defense against virus.
AGO5: binds with siRNA, vsiRNAs[3]
AGO6: binds with hc-siRNAs, siRNAs, ta-siRNAs and involved in DNA methylation
AGO7: binds with miR390 for ta-siRNA biogenesis
AGO9: binds with siRNAs
AGO10: binds with miR165, miR166, miR175, vsiRNAs
AGO3 and AtAGO8: Not characterized.
[1] ta-siRNAs (trans-acting small interfering RNAs)
[2] hc-siRNAs (heterochromatic siRNAs)
[3] vsiRNAs (virus-derived siRNAs)
- Judge, A.D., Sood, V., Shaw, J.R., Fang, D., McClintock, K. and MacLachlan, I. (2005) Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature biotechnology, 23, 457-462.
- Hornung, V., Guenthner-Biller, M., Bourquin, C., Ablasser, A., Schlee, M., Uematsu, S., Noronha, A., Manoharan, M., Akira, S., de Fougerolles, A. et al. (2005) Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nature medicine, 11, 263-270.
- Fedorov, Y., Anderson, E.M., Birmingham, A., Reynolds, A., Karpilow, J., Robinson, K., Leake, D., Marshall, W.S. and Khvorova, A. (2006) Off-target effects by siRNA can induce toxic phenotype. RNA, 12, 1188-1196.
- Armstrong, M.E., Gantier, M., Li, L., Chung, W.Y., McCann, A., Baugh, J.A. and Donnelly, S.C. (2008) Small interfering RNAs induce macrophage migration inhibitory factor production and proliferation in breast cancer cells via a double-stranded RNA-dependent protein kinase-dependent mechanism. Journal of immunology, 180, 7125-7133.
- Sledz, C.A., Holko, M., de Veer, M.J., Silverman, R.H. and Williams, B.R. (2003) Activation of the interferon system by short-interfering RNAs. Nat Cell Biol, 5, 834-839.
- McAllister, C.S. and Samuel, C.E. (2009) The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. The Journal of biological chemistry, 284, 1644-1651.
- Yu, Z., Teng, X. and Bonini, N.M. (2011) Triplet repeat-derived siRNAs enhance RNA-mediated toxicity in a Drosophila model for myotonic dystrophy. PLoS genetics, 7, e1001340.
- Lawlor, K.T., O'Keefe, L.V., Samaraweera, S.E., van Eyk, C.L. and Richards, R.I. (2012) Ubiquitous expression of CUG or CAG trinucleotide repeat RNA causes common morphological defects in a Drosophila model of RNA-mediated pathology. PloS one, 7, e38516.
- Sofola, O.A., Jin, P., Botas, J. and Nelson, D.L. (2007) Argonaute-2-dependent rescue of a Drosophila model of FXTAS by FRAXE premutation repeat. Human molecular genetics, 16, 2326-2332.
- Krzyzosiak, W.J., Sobczak, K., Wojciechowska, M., Fiszer, A., Mykowska, A. and Kozlowski, P. (2012) Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic acids research, 40, 11-26.
- Ahmed, F., Benedito, V.A. and Zhao, P.X. (2011) Mining functional elements in messenger RNAs: overview, challenges, and perspectives. Frontiers in Plant Science, 2.
- Dai, X. and Zhao, P.X. (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res, 39 Suppl 2, W155-159.
- Ahmed, F. and Raghava, G.P.S. (2011) Designing of Highly Effective Complementary and Mismatch siRNAs for Silencing a Gene. PLoS ONE, 6, e23443.
- Tafer, H., Ameres, S.L., Obernosterer, G., Gebeshuber, C.A., Schroeder, R., Martinez, J. and Hofacker, I.L. (2008) The impact of target site accessibility on the design of effective siRNAs. Nat Biotechnol, 26, 578-583.
- Muckstein, U., Tafer, H., Hackermuller, J., Bernhart, S.H., Stadler, P.F. and Hofacker, I.L. (2006) Thermodynamics of RNA-RNA binding. Bioinformatics, 22, 1177-1182.
- Fang, X. and Y. Qi (2016). "RNAi in Plants: An Argonaute-Centered View." Plant Cell 28(2): 272-285.
- Mi, S., T. Cai, Y. Hu, Y. Chen, E. Hodges, F. Ni, L. Wu, S. Li, H. Zhou, C. Long, S. Chen, G. J. Hannon and Y. Qi (2008). "Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5' terminal nucleotide." Cell 133(1): 116-127.