|
MET-COFEI:
A Novel Gas Chromatography-Mass Spectrometry Data Processing Platform for Metabolite Compound Feature Extraction and Identification
|
|
|
||
|
| Location: Introduction | |
|
GC-MS and LC-MS are complementary technologies that have been widely used in MS-based metabolomics studies. Compared with LC-MS, GC-MS is especially well suited for metabolites that are volatile and do not ionize well by LC-MS techniques. GC-MS provides high chromatograph resolution and permits separation of structurally similar compound that would be difficult to separate by LC. Additionally, the extracted pure spectrum not an individual peak for one compound can be reproduced with high quality but less affected by experiment reasons, which make GC-MS have the capability to identify metabolites by searching against some GC-MS spectra library. However, hundreds of metabolites could exist in a biological sample together with noise and contaminant interference, which make it still a challenge to extract the pure spectra for the following component identification. Herein, we present a novel GC-MS data processing and analysis platform entitled as MET-COFEI(METabolite COmpound Feature Extraction and Identification), which can be used in GC-MS based metabolomics study. MET-COFEI is aiming to extract the acute EIC(also called mass trace) by object tracing method, and then the pure compound related spectra by detecting and clustering the meaningful chromatograph peak features based on the retention time and peak shape criteria, and then identify the metabolite by searching against some spectra library. MET-COFEI has been implemented as a seamless pipeline for data analysis and suit for high-throughout metabolomics profiling. Fig.1 gives the illustration of MET-COFEI workflow, at first, 5 meaningful mass traces are extracted and outlined in m/z-retention time domain, then the chromatograph peaks are extracted and further grouped according to peak retention time and shape similarity, finally grouped into a peak list. The right spectrums illustrate how a mixed and noisy spectrum can be filtered/purified into a meaningful authentic compound related spectrum. The output results of MET-COFEI are the peak list tables that have been clustered and grouped into different component groups by unique Group_ID, by which, a compound associated pure spectrum can be constructed and searched against an open GC-MS library. If found any, the compound’s chemical name of the top matched library searching result will be attached to the column of Compound_Name. Otherwise, ’Unknown’ will be filled. 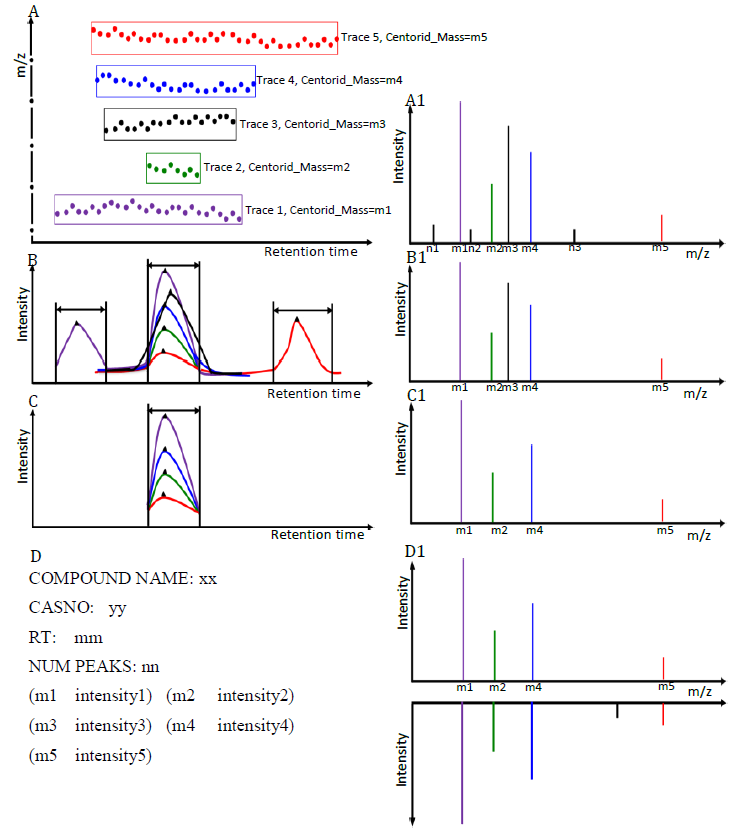 Fig. 1 Illustration of MET-COFEI workflow METCOFEI can be summarized as three integrative modules: Compound Feature Extraction, Compound Identification and Compound Alignment(see Fig.2). Compound feature extraction module aims to extract individual peaks that corresponding to compound’s fragments from each inputting GC/MS raw data. Compound identification module aims to cluster/group peaks based on peak shapes, retention time, and then construct the pure spectrum for each potential compound(also called component in some papers), and further to identify the compound by searching against an open GC-MS library. Compound alignment module is aiming to align the same compound based on the similarity between the two compound-associated pure spectrums across different samples.. 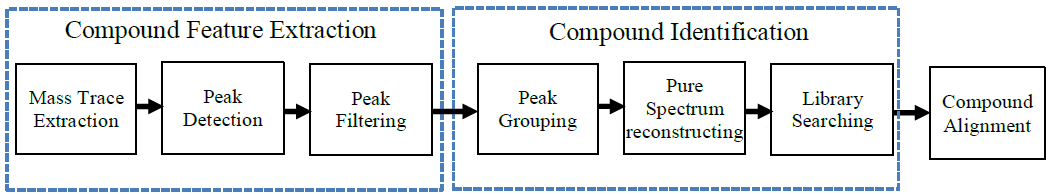 Fig. 2 Three integrative key modules in MET-COFEI The basic architecture of MET-COFEI is depicted in Fig.3. It mainly includes a core and user interface. The core of MET-COFEI(entitled as MET-COFEI Engine) mainly includes several data processing modules and several assisting modules, which is written standard C++ using Standard Template Libraries (STL). The user interface mainly includes interactive parameter configuration and visualization. Additionally, a wrapper is written in CLI/C++, which acts as a bridge between the front-end visualization and the back-end processing. All of the codes are developed and compiled in Microsoft Visual Studio Ultimate 2012. 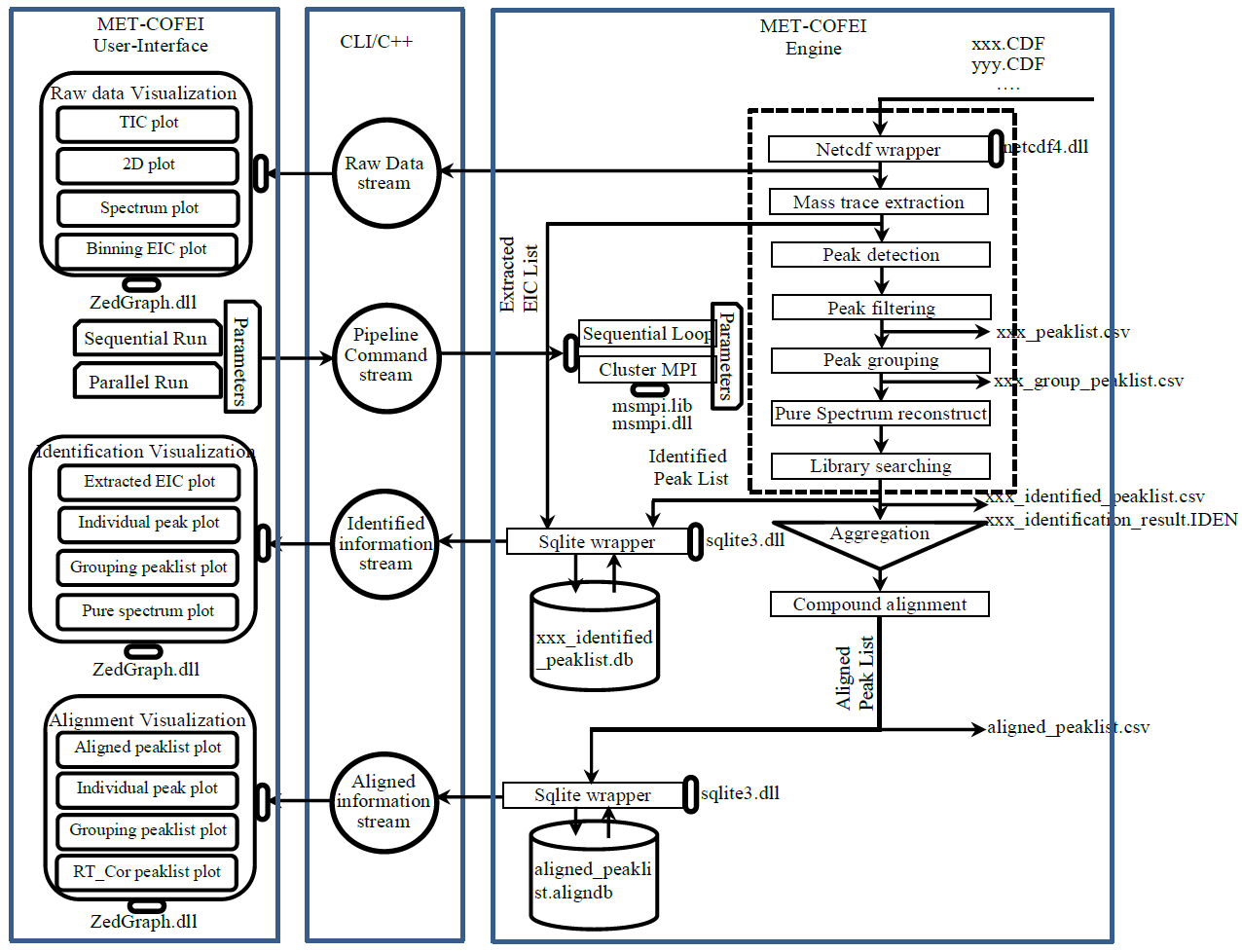 Fig. 3 Implementation architecture of MET-COFEI Using MET-COFEI, the user can visualize the selected sample (.CDF raw data) and process the loaded data samples at batch or parallel mode(Fig.4), once the parameters are configured(Fig.5) and loaded. Additionally, a GC-MS library file(.msl/.MSP format) is needed to be specified for compound identification(Fig.6). 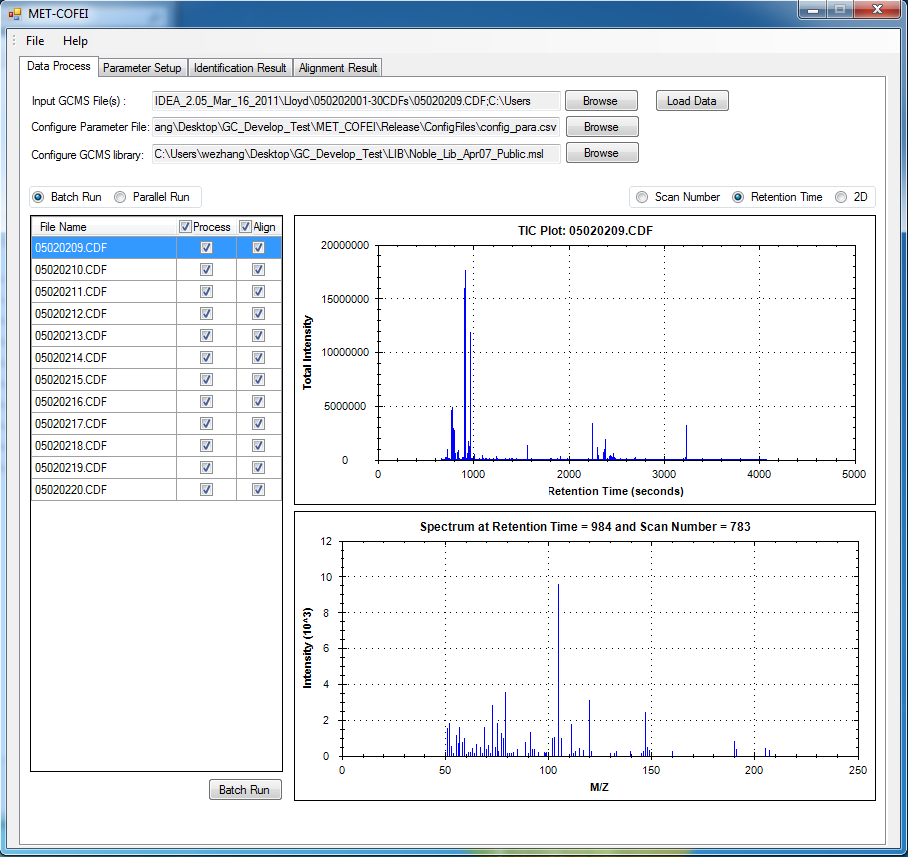 Fig. 4 Raw data visualization and pipeline processing by MET-COFEI 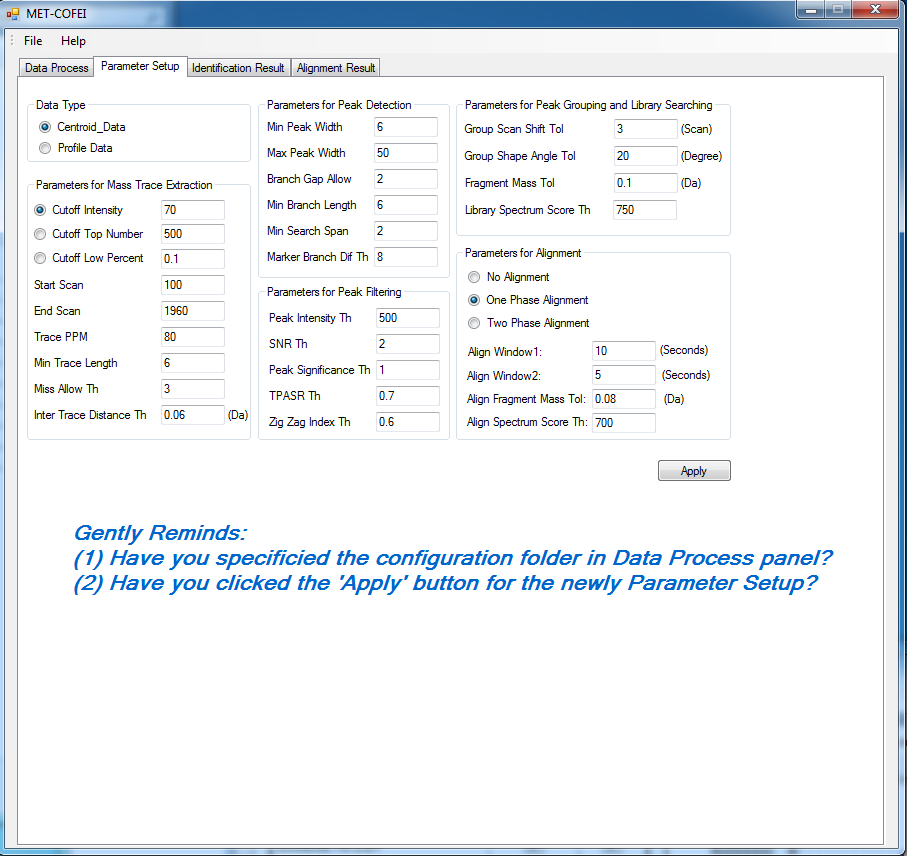 Fig. 5 MET-COFEI parameter configuration 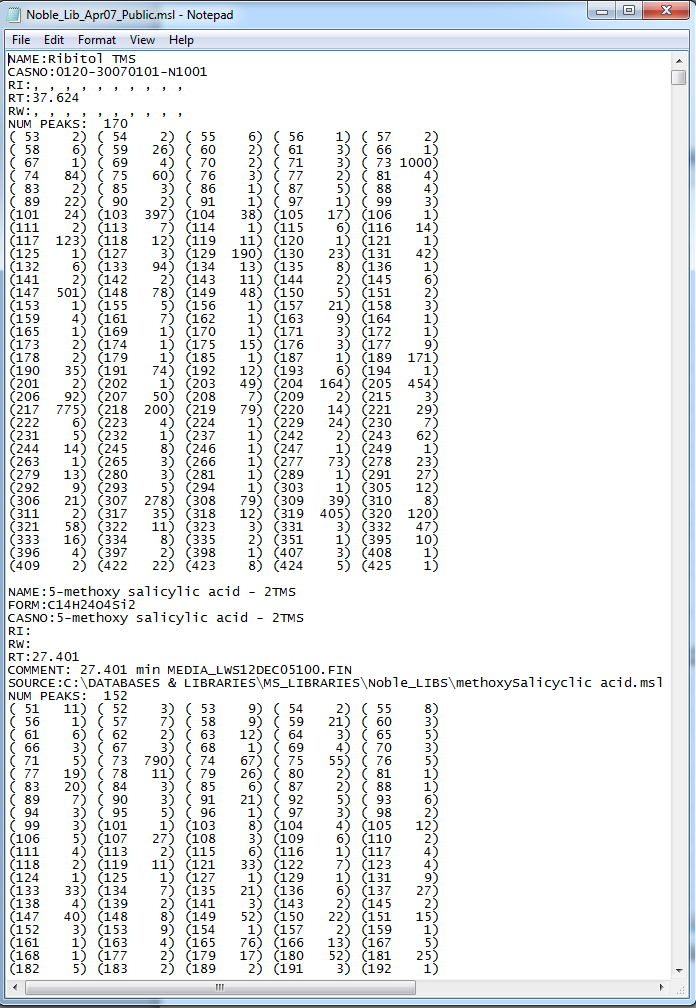 Fig. 6 Snapshot of a GC-MS library file that can be used in MET-COFEI Once the analysis for one sample finished, MET-COFEI, utilizing SQLite technique, will output a database file named as xxx.db. User can visualize the detail of grouping and library searching based identification results. Fig.7 give screenshots of the associated peaks for the same Group_ID. The peak list with the same Group_ID are close in peak shape and retention time. The constructed pure spectrum with the same Group_ID can be exported as a txt file. The final identification result file(.IDEN file) recorded the identification results for each extracted pure spectrum against the specific GC-MS library(Fig.8). The matching score is based the similarity between the extracted spectrum and library spectrum. 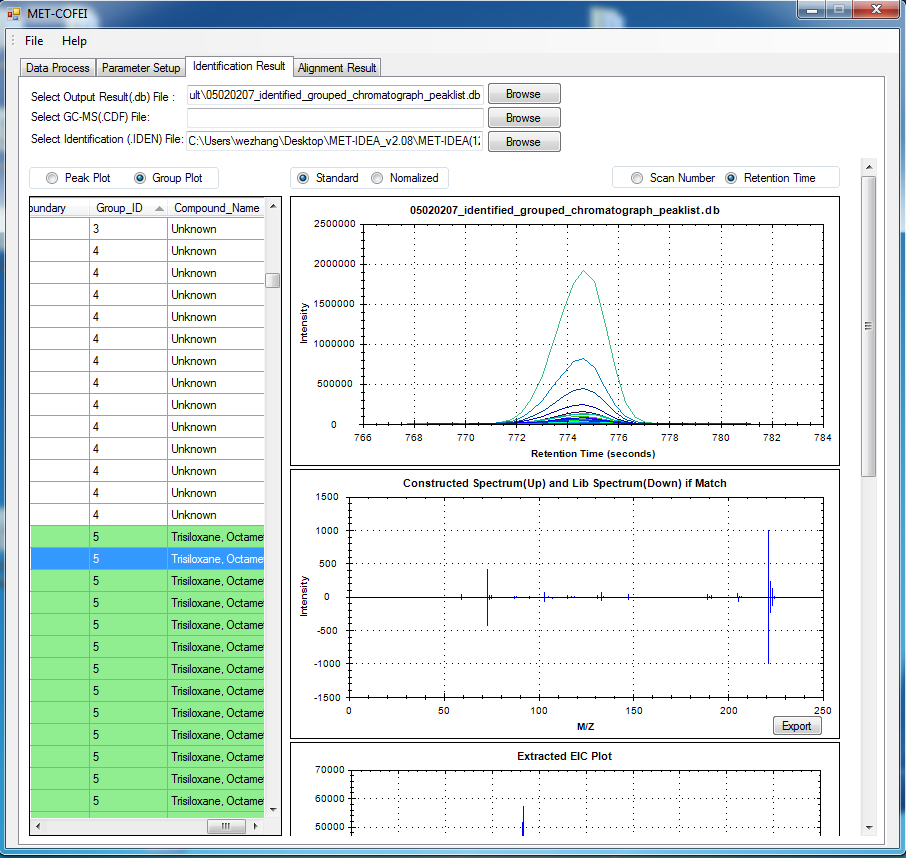 Fig. 7 Visualization of MET-COFEI’s Identification results for the grouped peaks with the same Group_ID 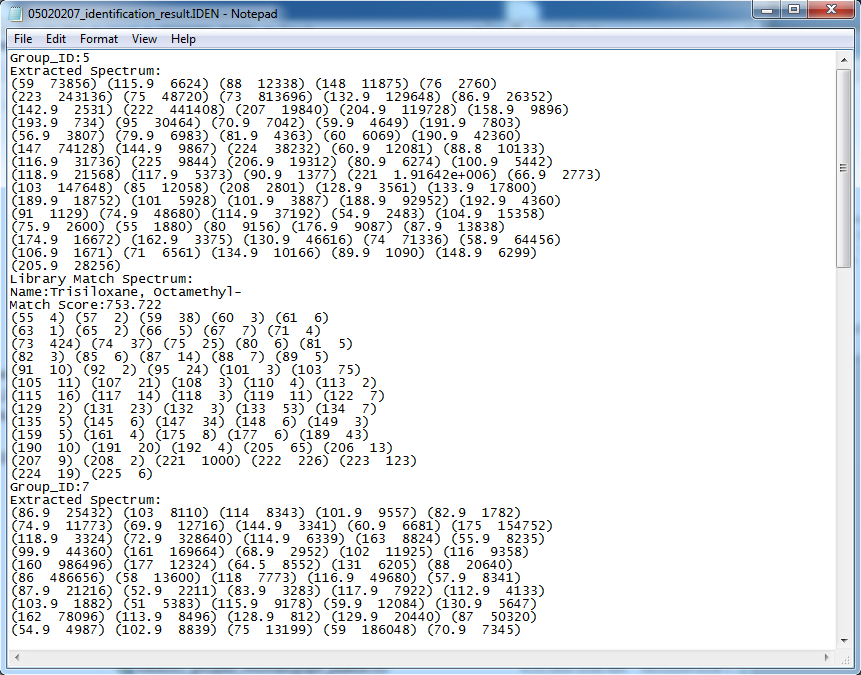 Fig. 8 Snapshot of Identification result file based on the library searching for each extracted pure spectrum against the user specific GC-MS library Once the alignment across samples finished, MET-COFEI, utilizing SQLite technique, will output a database file named as aligned_identified_grouped_chromatograph_peaklist.aligndb. User can visualize the detail of alignment results. Fig.9 give screenshots of the associated peaks for the same Align_ID at the corrected retention time. The identified compound associated peaks can be aligned together if the spectrum similarity score and retention time fall in the user specific tolerance. 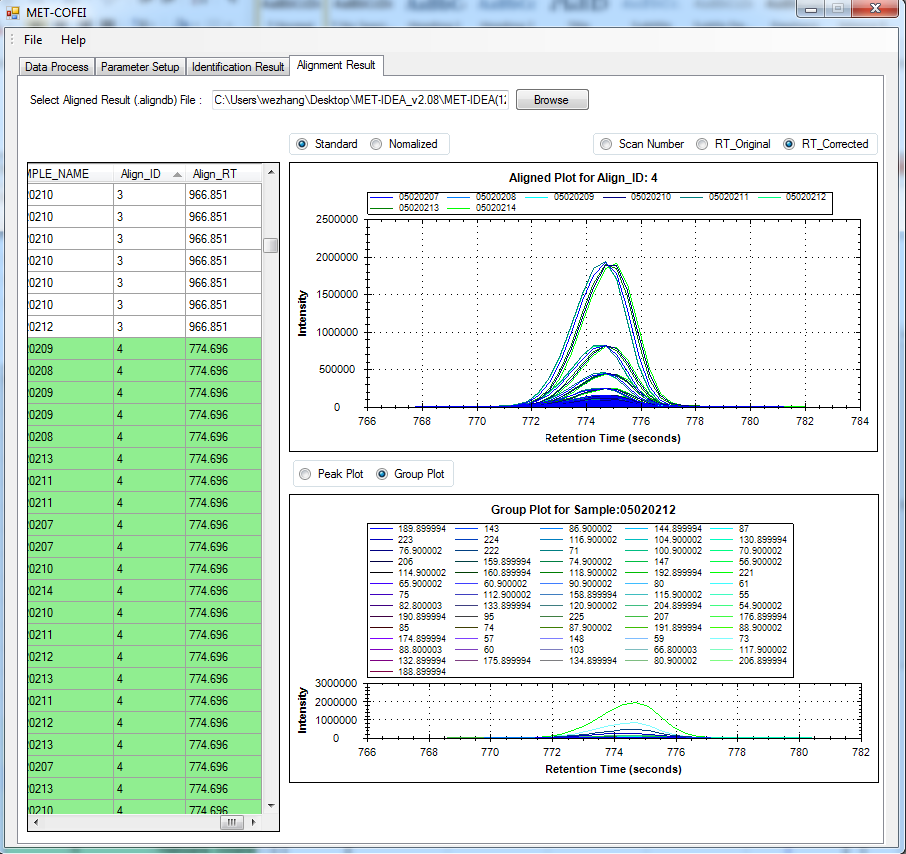 Fig. 9 Visualization of MET-COFEI’s Alignment results for the aligned peaks with the same Align_ID For any questions or suggestions about MET-COFEI, pls feel free to contact: bioinfo AT noble.org |
| © Copyright 2013, the Samuel Roberts Noble Foundation, Inc. Developed by The Zhao Lab |