|
Case Study 1 (258 rice accessions X 1,619 SNP BINs) If you use these data for publication, please use the following citation: Xu, Shizhong. "Genetic mapping and genomic selection using recombination breakpoint data." Genetics 195.3 (2013): 1103-1115. 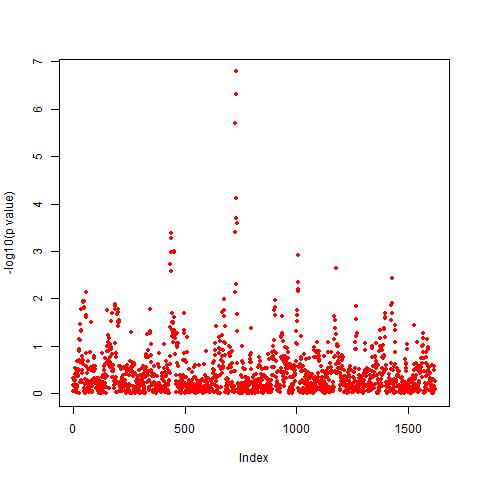
|
|
| Data download: | Genotypic data; Phenotypic data; Kinship matrix |
|
Case Study 2 (220 Medicago truncatula accessions X 1,810,466 SNPs) If you use these data for publication, please use the following citation: Kang, Yun, et al. "Genome‐wide association of drought‐related and biomass traits with HapMap SNPs in Medicago truncatula." Plant, cell & environment 38.10 (2015): 1997-2011. 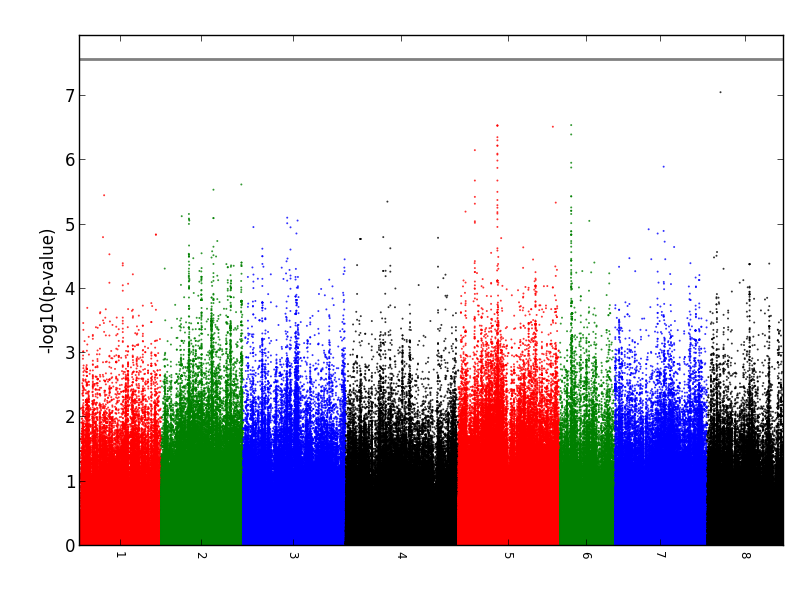
|
|||
| Data download: | Genotypic data; Phenotypic data; Kinship matrix; SNP map | ||
|
Case Study 3 (302 Maize accessions X 62,077 SNPs) If you use these data for publication, please use the following citation: Sanchez, Darlene L., et al. "Genome-wide association studies of doubled haploid exotic introgression lines for root system architecture traits in maize (Zea mays L.)." Plant Science 268 (2018): 30-38. 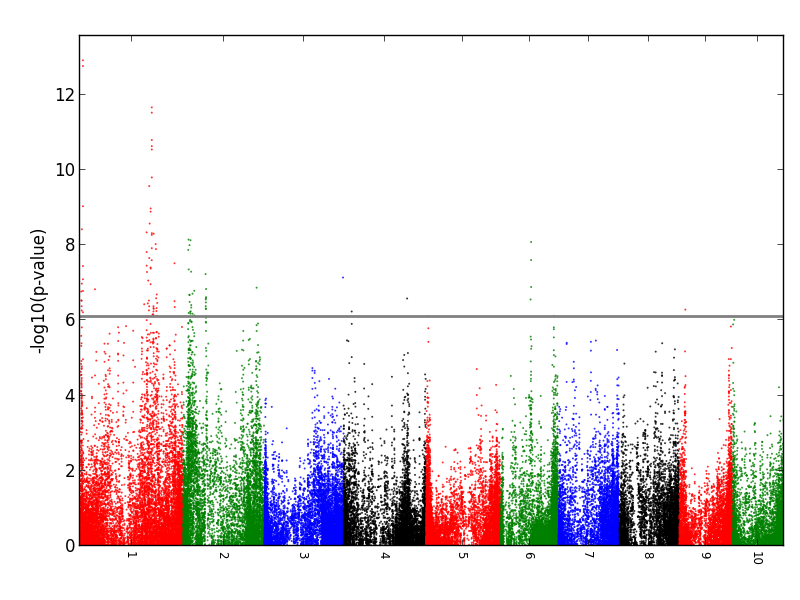
|
|||
| Data download: | Genotypic data; Phenotypic data; SNP map | ||