Data normalization is an important step before gene association network construction. To assist with data normalization required by the GPLEXUS, user use the following three options to normalize their Affymetrix GeneChip data based on the Robust Multi-array Average (RMA) method (log2 scale is not necessary) :
- Option1: Use our online normalization tool
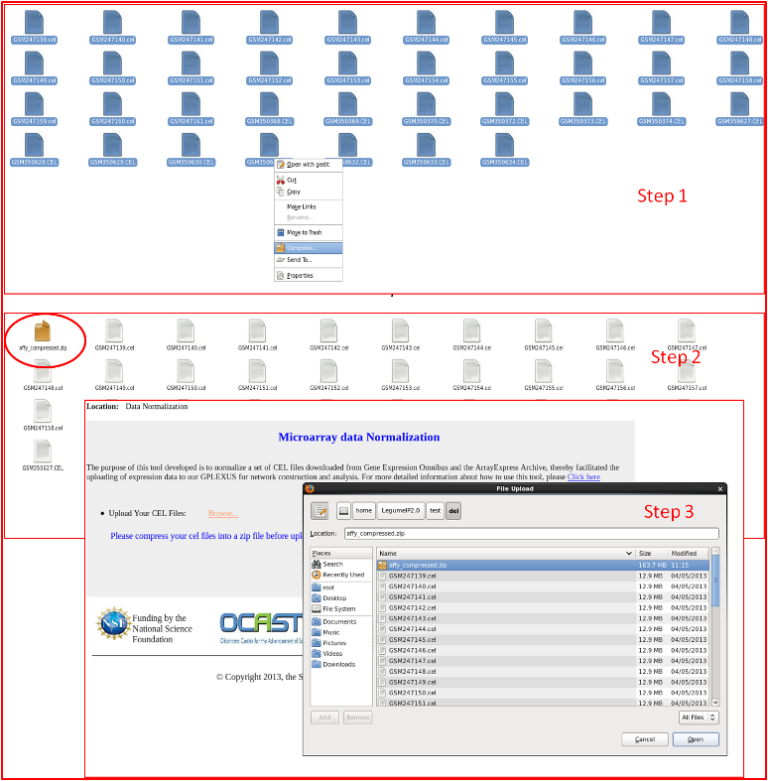
User needs to upload a zipped file containing the Affymetrix CEL-files to our webserver to perform normalization.
- Option2: Use "Affy" module in the R package
If a user has more than several hundreds of CEL files, we recommend using the "affy" R package to perform data normalization on local computers. Below are steps how to normalize affymetrix data and save it into a TAB-delimited txt file.
1. Install R environment and "affy" R package;
2.Run the following scripts:
|
library(affy)
setwd(“/tmp/affymetrix”)
Data <- ReadAffy()
eset <- rma(Data)
write.exprs(eset,file="rma.txt")
|
Please note that the affymetrix expression data will be read from and saved into the folder “/tmp/affymetrix/”. User may change it to any other directory where the affymetrix data is located. The normalized the table-delimited expression data file will be saved in “rma.txt”.
- Option3: Install and run RMA software under windows environment




