|
|
|
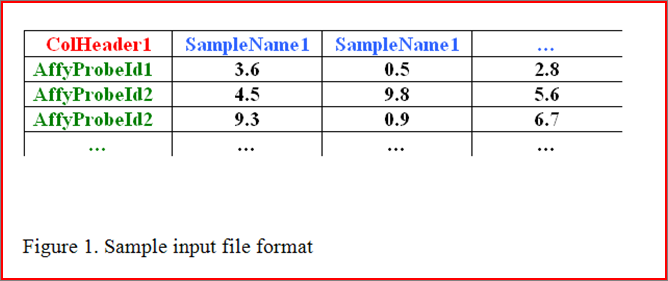
Data formats allowed by GPLEXUS:
Our GPLEXUS only reads TAB-delimited text files in a particular format, described below. Such files can be created and exported in any standard spreadsheet program, such as Microsoft Excel
|
Mutual Information Estimation Method
GPLEXUS provides three methods to estimate Mutual information. One method estimating MI using the transformation formula of Spearman-correlation method, the second method is based on B-Spline functional and the third method is based on Gaussian kernel estimator method as in the ARACNE paper;
In terms of their accuracy, their is no big difference between these three methods. However, Spearman correlation-based method is far faster than Gaussian kernel-based method.
So we suggest users use the formula of Spearman-correlation method to estimate mutual information for massive datasets.
|
The Output File
GPLEXUS will provide three files for downloading. The 'Network' column will output a list of gene interactions in a TAB-delimited text files and such format is very easy to be loaded into Cytoscape Software;
The "Probset/Gene Connectivity" column will output a text file including gene and the number of interaction with this gene in a descending order;
The "Module" column will output a compressed file in the zip format. It included multiple files names as "Module_***" that are used to store gene interactions for every module. Another file named as "Module_gene.txt" will be used to store gene list for every module, and every line in this file will be used to store the gene list for every module. For example, the first line denote as the gene locate in the first module and second line denote as genes located in the second module.
The Network Result
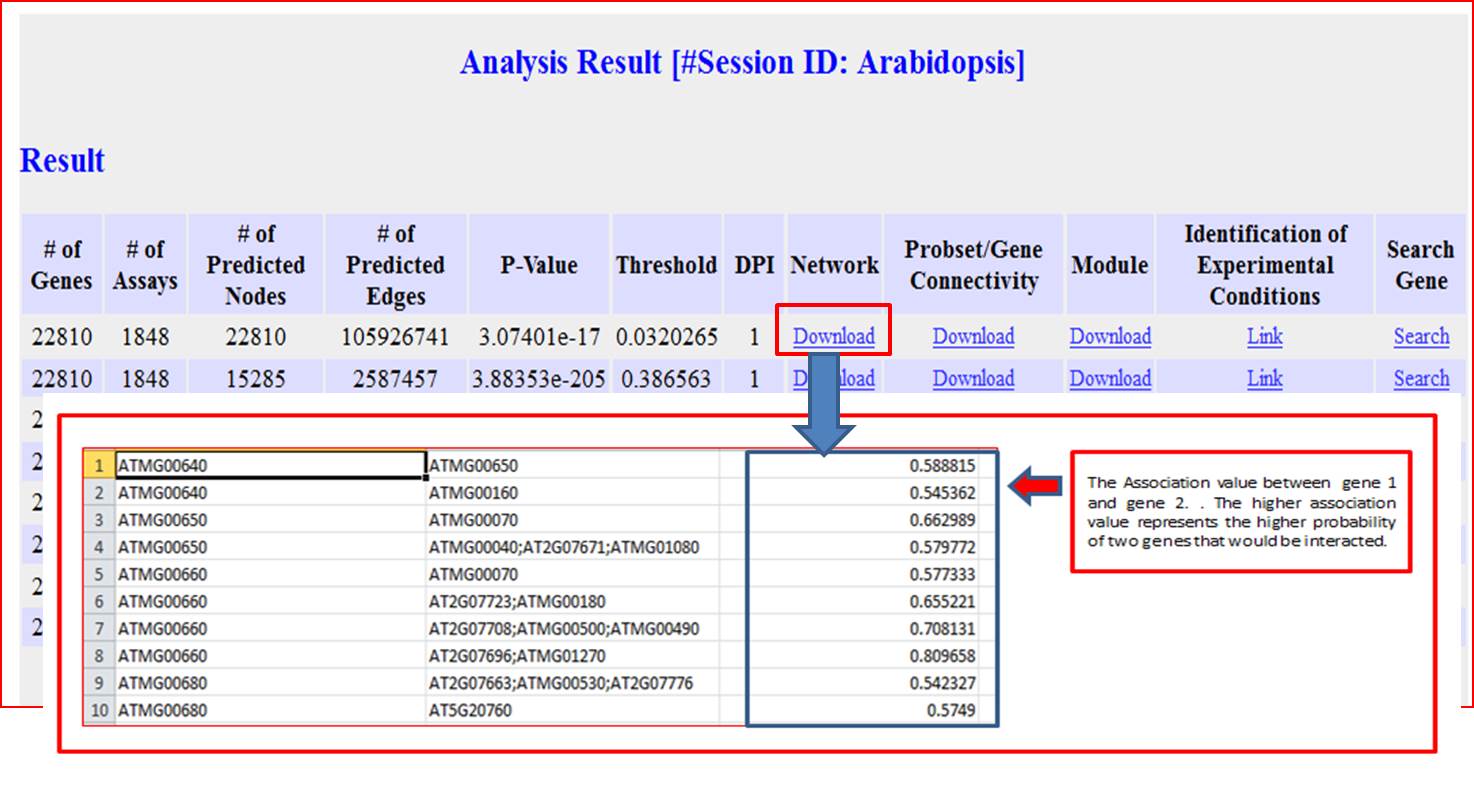
The Parameter Setting
Mi Threshold: The GPLEXUS uses mutual information(MI),in information theoretical measure to compute the association between pairs of genes (I.E. an MI score of 0 between two genes implies they are independent of each other ,and the higher MI score indicate the probability of interaction between two genes is higher).
DPI tolerance: The DPI(data processing inequality) tolerance is used to reduce the number of genes which do not interact directly but were missed in the MI thresholding step. Lower DPI value will be used to reduce the number of potential indirect edges.
If you are interested in constructing higher probability network (it means smaller number of genes and edges in the network), you should use a higher MI threshold and lower DPI value setting.
Identification of Experiment-specific condition
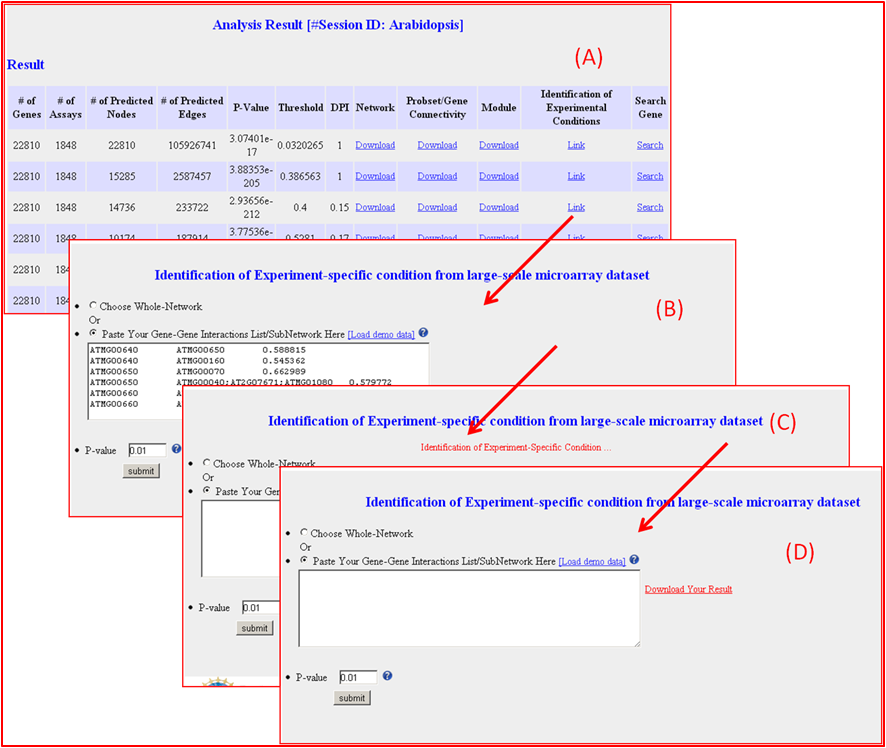
Search and Visualization
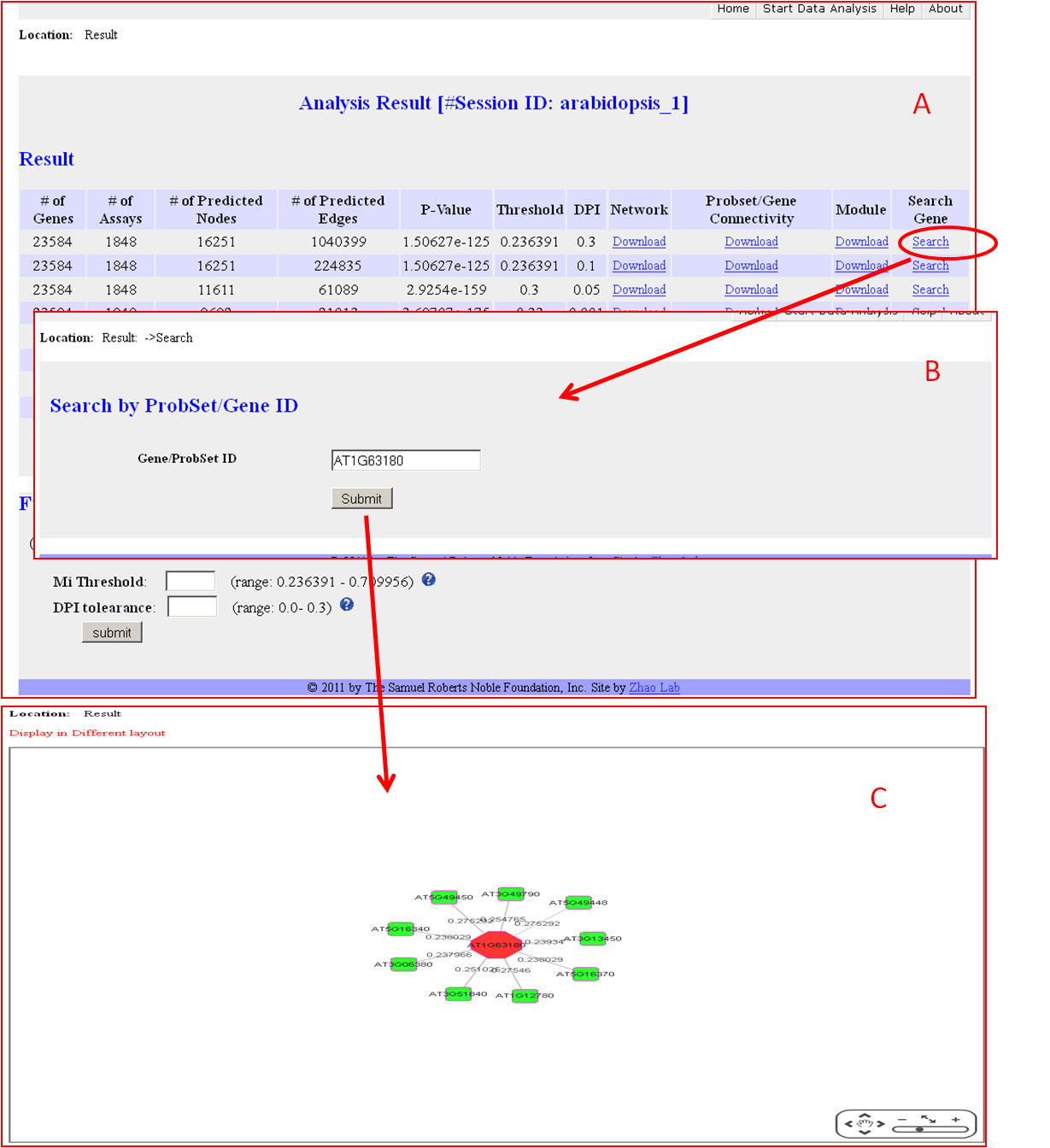
Funding:
- This work was supported by National Science Foundation (Grant ABI-0960897) and the Samuel Roberts Noble Foundation.
Contact:
- To contact us, please write to: bioinfo AT noble DOT org
|
 |
Funding by the National Science Foundation |
 |
Funding by the Oklahoma Center for the Advancement of Science & Technology |
 |
Additional funding by the Samuel Roberts Noble Foundation |
|
© Copyright 2013, the Samuel Roberts Noble Foundation, Inc. Site by Zhao Lab
|
|







